Researchers at the Max Planck Institute for Chemical Ecology in Jena and their colleagues from the Czech Academy of Sciences in Prague have developed a new method to quickly and reliably detect metabolites, such as sugars, fatty acids, amino acids and other organic substances from plant or animal tissue samples. One drop of blood -- less than one micro liter -- is sufficient to identify certain blood related metabolites.
Background:Mass spectrometry is an analytical technique used to elucidate the
molecular composition and structure of chemical compounds. In the last
two decades mass spectrometry found vast applications in biology,
especially for analyzing of large biomolecules. Matrix-Assisted Laser
Desorption/Ionization (MALDI), wherein bio-molecules (e.g. proteins)
are co-crystallized with a chemical substance called a matrix
subsequently irradiated with a laser leads to the formation of protein
ions which can be analyzed and detected.
However, matrices
used in the MALDI technique have a substantial disadvantage: the laser
beam not only forms ions from the substances of interest; it also forms
low-mass ions (<500 Da) originating from the matrix. "Because of
these small interfering ions we were not able to analyze small
molecules that play crucial roles in the metabolism of organisms,"
explains Aleš Svatoš, head of the mass spectrometry/proteomics research
group at the Max Planck Institute. "The ions that originated from
conventional matrices were like a haystack in which we wanted to find a
few and important needles." Therefore the MALDI technique found only
limited application in the field of "metabolomics".
The new technique:
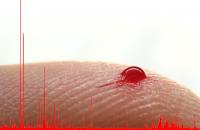 |
Image: One drop of blood is sufficient for detailed metabolomic analysis by MAILD mass spectrometry.
Credit: MPI for Chemical Ecology, Aleš Svatoš
|
The new technique, called MAILD, is based on classical mass
spectrometry (MALDI-TOF/MS) and enables researchers to measure a large
number of metabolites in biological samples, opening doors for targeted
and high-throughput metabolomics. Because of its versatile
applications, also in medical diagnostics, the invention is protected
by patent.
Instead of improving the search for the "needles", i.e. metabolites
such as sugars, fatty acids, amino acids, and other organic acids, the
scientists began to alter the matrices with which the samples were
applied so that no more interfering matrix-related ions were generated.
In other words: they tried to remove the haystack to make the needles
visible. The researchers succeeded with the help of physical and
organic chemistry, based on the Brřnsted–Lowry acid-base theory, and
formulated conditions for rational selection of matrices that did not
generate interfering ions but provided rich mass spectra of particular
kinds of metabolites in real samples.
With the new
experimental protocols they called "Matrix-Assisted Ionization/Laser
Desorption – MAILD", the scientists were able to quickly and reliably
determine more than 100 different analytes from single and small-sized
samples. "The analysis of a very small plant leaf sample from Arabidopsis thaliana,
in fact a circle area with a radius of just about 0.5 millimeter,
revealed over a hundred analyte peaks, among which 46 metabolites could
be identified. Interestingly, among them were eight of a total of
eleven intermediates of the citric acid cycle, which is vital for most
organisms," says Rohit Shroff, a native of India, who was a PhD student
at the "International Max Planck Research School" and conducted the
experiments.
The new MAILD method allows measurements from
diverse biological and medical materials. Apart from plant and insect
samples the scientists also studied a clinical sample: they were able
to determine a wide range of blood-specific organic acids in one drop
of human blood, smaller than a micro liter. In medical diagnostics such
measurements are still conducted with intricate methods. If the
scientists succeed in not only identifying, but also quantifying the
metabolites, MAILD could develop into a fast method for medical and
biological diagnostics.
Source: Max Planck Institute for Chemical Ecology
 The original publication
The original publication
 Rohit Shroff, Lubomír Rulíšek, Jan Doubský, Aleš Svatoš, Acid-base-driven matrix-assisted mass spectrometry for targeted
metabolomics, Proceedings of the National Academy of Sciences USA, 106/25 (2009) 10092-10096. DOI: 10.1073/pnas.0900914106.
Rohit Shroff, Lubomír Rulíšek, Jan Doubský, Aleš Svatoš, Acid-base-driven matrix-assisted mass spectrometry for targeted
metabolomics, Proceedings of the National Academy of Sciences USA, 106/25 (2009) 10092-10096. DOI: 10.1073/pnas.0900914106.
 Related EVISA Resources
Related EVISA Resources Brief Summary: Tools for elemental speciation
Brief Summary: Tools for elemental speciation Brief Summary: ICP-MS - A versatile detection system for speciation analysis
Brief Summary: ICP-MS - A versatile detection system for speciation analysis Brief Summary: LC-ICP-MS - The most often used hyphenated system for speciation analysis
Brief Summary: LC-ICP-MS - The most often used hyphenated system for speciation analysis Brief Summary: GC-ICP-MS: A very sensitive hyphenated system for speciation analysis
Brief Summary: GC-ICP-MS: A very sensitive hyphenated system for speciation analysis Brief Summary: CE-ICP-MS for speciation analysis
Brief Summary: CE-ICP-MS for speciation analysis Brief Summary: ESI-MS: The tool for the identification of species
Brief Summary: ESI-MS: The tool for the identification of species  Related EVISA News
Related EVISA News January 15, 2008: Species-specific isotope dilution analysis has been adopted as an official method under US legislation
January 15, 2008: Species-specific isotope dilution analysis has been adopted as an official method under US legislation  November 8, 2006: Double spiking species-specific isotope dilution result calculations simplified
November 8, 2006: Double spiking species-specific isotope dilution result calculations simplifiedlast time modified: May 21, 2024