More mercury species than previously thought can be converted by bacteria to methylmercury and thus be accumulated along the food chain.
The discovery, published last Sunday in Nature Geosciences, is adding another piece of the puzzle desribing the mercury cycling in the environment and will hopefully help to elucidate the challenges associated with mercury pollution and its cleanup.
Background:
Mercury is a toxin that spreads around the globe mainly through the burning of coal, other industrial uses, and natural processes such as volcanic eruptions, and various forms of mercury are widely found in sediments and water. Methylmercury bioaccumulates in aquatic food chains, especially in large fish.
Earlier this year, a multidisciplinary team of researchers at Oak Ridge National Laboratory discovered two key genes that are essential for microbes to convert oxidized mercury to methylmercury, a neurotoxin that can penetrate skin and at high doses affect brain and muscle tissue, causing paralysis and brain damage (see the EVISA News below).
The discovery of how methylmercury is formed answered a question that had stumped scientists for decades, and the findings published this week build on that breakthrough.
Most mercury researchers have believed that microbes could not convert elemental mercury -- which is volatile and relatively inert -- into methylmercury. Instead of becoming more toxic, they reasoned that elemental mercury would bubble out of water and dissipate. That offered a solution for oxidized mercury, which dissolves in water. By converting oxidized mercury into elemental mercury, they hoped to eliminate the threat of methylmercury contamination in water systems.
The new study:
ORNL's study and a parallel study reported by Rutgers University,
however, suggest that elemental mercury is also susceptible to bacterial
manipulation, a finding that makes environmental cleanup more
challenging.
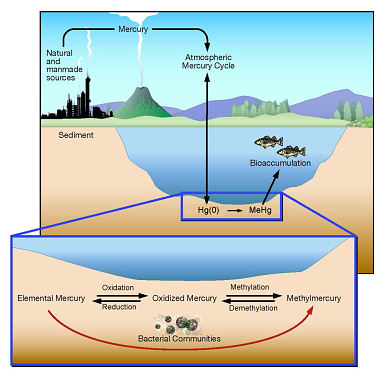
Figure: ORNL researchers are learning more about the microbial
processes that convert elemental mercury into methylmercury.
Credit: Department of Energy's Oak Ridge National Laboratory
|
"Communities of microorganisms can work together in environments that lack oxygen to convert elemental mercury to methylmercury," study leader Baohua Gu said. "Some bacteria remove electrons from elemental mercury to create oxidized mercury, while others add a methyl group to produce methylmercury."
The fight against mercury pollution involves scientists with expertise in chemistry, computational biology, microbiology, neutron science, biochemistry and bacterial genetics. Other ORNL efforts are focusing on when, where and why bacteria are producing methylmercury.
"Our research allows us to understand generally where and how bacteria might produce methylmercury so that we can target those areas in the future," said ORNL's Liyuan Liang, a co-author and director of the DOE-funded mercury research program. "We are trying to understand the process of microbial mercury methylation. Once we understand the process, we can begin to form solutions to combat mercury pollution."
Source: Adapted from
DOE/Oak Ridge National Laboratory  The original study
The original study:

Haiyan Hu, Hui Lin, Wang Zheng, Stephen Tomanicek, Alexander Johs, Dwayne Elias, Liyuan Liang and Baohua Gu,
Oxidation and methylation of dissolved elemental mercury by anaerobic bacteria, Nature Geoscience, 6/9 (2013) 751-754.
doi: 10.1038/ngeo1894
 Related studies
Related studies:

Matthew J. Colombo, Juyoung Ha, John R. Reinfelder, Tamar Barkay, Nathan Yee,
Anaerobic oxidation of Hg(0) and methylmercury formation by Desulfovibrio desulfuricans ND132, Geochim. Cosmochim. Acta, 112 (2013) 166–177.
doi: 10.1016/j.gca.2013.03.001
Jeffra K. Schaefer, Sara S. Rocks, Wang Zheng, Liyuan Liang, Baohua Gu, François M. M. Morel,
Active transport, substrate specificity, and methylation of Hg(II) in anaerobic bacteria, Proc. Natl. Acad. Sci. U.S.A., 108 (2011) 8714–8719.
doi: 10.1073/pnas.1105781108
Cynthia C. Gilmour, Dwayne A. Elias, Amy M. Kucken, Steven D. Brown, Anthony V. Palumbo, Christopher W. Schadt, Judy D. Wall,
Sulfate-Reducing Bacterium Desulfovibrio desulfuricans ND132 as a Model for Understanding Bacterial Mercury Methylation, Appl. Environ. Microbiol., 77/12 (2011) 3938-3951.
doi:10.1128/AEM.02993-10 
Jeffra K. Schaefer, François M. M. Morel,
High methylation rates of mercury bound to cysteine by Geobacter sulfurreducens, Nature Geosci., 2/2 (2009) 123-126.
doi: 10.1038/ngeo412
Ariane Bouffard,
Marc Amyot,
Importance of elemental mercury in lake sediments, Chemosphere, 74 (2009) 1098–1103.
doi: 10.1016/j.chemosphere.2008.10.045 Marc Amyot
Marc Amyot, Francois M.M. Morel, Parisa A. Ariya,
Dark Oxidation of Dissolved and Liquid Elemental Mercury in Aquatic Environments, Environ. Sci. Technol., 2005, 39, 110-114.
doi: 10.1021/es035444k
Jenny Ayla Jay, Karen J. Murray, Cynthia C. Gilmour, Robert P. Mason, Francois M.M. Morel, A. Lynn Roberts, Harold F. Hemond,
Mercury Methylation by Desulfovibrio desulfuricans ND132 in the Presence of Polysulfides, Appl. Environ. Microbiol., 68 (2002) 5741-5745.
DOI: 10.1128/AEM.68.11.5741-5745.2002
S.D. Siciliano, N.J. D'Driscoll, D.R.S. Lean,
Microbial Reduction and Oxidation of Mercury in Freshwater Lakes, Environ. Sci. Technol. 2002, 36, 3064-3068.
doi: 10.1021/es010774v Related EVISA Resources
Related EVISA Resources Link database: Mercury exposure through the diet
Link database: Mercury exposure through the diet Link database: Mercury and human health
Link database: Mercury and human health Link database: Environmental cycling of mercury
Link database: Environmental cycling of mercury Link database: Toxicity of Organo-mercury compounds
Link database: Toxicity of Organo-mercury compounds Link database: Research projects related to organo-mercury compounds
Link database: Research projects related to organo-mercury compounds Related EVISA News
Related EVISA News
 June 17, 2012: Factors Affecting Methylmercury Accumulation in the Food Chain
June 17, 2012: Factors Affecting Methylmercury Accumulation in the Food Chain  October 9, 2006: Linking atmospheric mercury to methylmercury in fish
October 9, 2006: Linking atmospheric mercury to methylmercury in fish  February 17, 2006: Study shows link between clear lakes and methylmercury contamination in fish
February 17, 2006: Study shows link between clear lakes and methylmercury contamination in fish last time modified: January 19, 2025