Researchers from the University of Münster (Germany) identified carbonic anhydrase as a mercury transporting protein in human blood
Background:
Organic mercury species such as Methylmercury and Ethylmercury are widely known to be highly toxic. Humans are exposed to Methylmercury mainly via consumption of marine food, especially of carnivorous fish species that have bioaccumulated methylmercury and biomagnified its concentration along the food-chain. Moreover, thimerosal (THI) is another mercury containing substance, which is used as a preservative in multidose ampullae of vaccines and other drugs. The antimicrobial and toxic effects of THI are based on its decomposition in aqueous media releasing a (mono)-ethylmercury cation (EtHg+). The molecular mechanisms of MeHg+ and EtHg+ toxicity and toxic kinetics are not fully understood and still are a topic of ongoing research. By entering the body, the organic mercury species are distributed via the blood stream to the different organs. Due to the high affinity of mercury towards sulfur, reactions with free thiol groups of peptides and proteins with different mercury species may occur. It is assumed that the interaction of mercury species with thiols plays an important role for mercury transport, overcoming certain barriers such as the blood/brain barrier and for their toxic mechanisms of action. Prior studies have shown that most of the mercury in blood is bound to the erythrocytes and one of the binding partners, namely hemoglobin, has been identified.
The new study:Researchers from the University of Münster now investigated whether further biomolecules interact with the organic mercury species. They concentrated their attention towards another highly abundant protein in blood, namely the carbonic anhydrase (CA). This enzyme catalyzes the reversible hydration of carbon dioxide and is found in all animals and photosynthesizing organisms. Using in vitro experiments under physiological conditions, the researchers indeed were able to identify the adduct formation between CA and the organic mercury species by means of LC/ESI-ToF-MS and LC/ICP-MS.
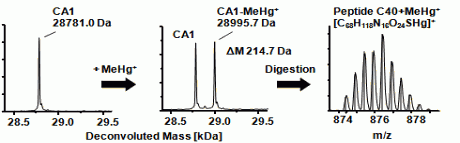
Figure 1: The adduct formation between CA1 and Methylmercury shows up in the ESI-MS spectrum by the addition of 214.7 DA. After enzymatic digestion of the CA, the methylmercury binding site can be identified by the Mercury isotopic pattern within the peptide-adduct.
Adduct formation between CA and mercury was found for both methylmercury as well as ethylmercury. The adduct could be identified by its exact mass as well as the isotope fingerprint of mercury. To specify the expected binding site, an enzymatic digest was performed. Trypsin and chymotrypsin were used in parallel for the digestion to obtain different peptide patterns. The tryptic digest of CA1 results in 24 specific peptides, in which the peptide T19 (#174–213) is the peptide featuring the only cysteine residue and consists of 40 amino acids. The digestion with chymotrypsin produced 45 fragments from CA1. The peptide C40, which contains the cysteine, consists of 14 amino acids (#210–223). In both digestion experiments, mercury binding was found at the only cysteine containing peptide. In this way it was verified that carbonic anhydrase, and not only hemoglobin, provides such binding site for organic mercury species and forms the related adducts. Since both proteins have a high abundance in blood, they both are expected to play a role in the
transport of mercury in blood.
 The original studies
The original studies

Jens Hogeback, Miriam Schwarzer,
Christoph A. Wehe,
Michael Sperling,
Uwe Karst,
Investigating the adduct formation of organic mercury species with carbonic anhydrase and hemoglobin from human red blood cell hemolysate by means of LC/ESI-TOF-MS and LC/ICP-MS, Metallomics, 8/1 (2015) 101-107.
DOI: 10.1039/c5mt00186b Homepage
of the research group of Prof. Karst at the Institute of Inorganic and
Analytical Chemistry of the University of Muenster (Germany)
Homepage
of the research group of Prof. Karst at the Institute of Inorganic and
Analytical Chemistry of the University of Muenster (Germany)
 Related Studies (newest first)
Related Studies (newest first)

S. Trümpler,
B. Meermann, S. Nowak,
W. Buscher,
U. Karst,
M. Sperling,
In vitro study of thimerosal reactions in human whole blood and plasma surrogate samples, J. Trace Elem. Med. Biol., 28 (2014) 125–130.
DOI: 10.1016/j.jtemb.2014.01.006
Z. Pedrero Zayas, L. Ouerdane, S. Mounicou,
R. Lobinski,
M. Monperrus,
D. Amouroux,
Hemoglobin as a major binding protein for methylmercury in white-sided dolphin liver, Anal. Bioanal. Chem., 406 (2013) 1121–1129.
DOI 10.1007/s00216-013-7274-6

D. J. Kutscher,
A. Sanz-Medel,
J. Bettmer,
Metallomics investigations on potential binding partners of methylmercury in tuna fish muscle tissue using complementary mass spectrometric techniques, Metallomics, 4 (2012) 807–813.
DOI: 10.1039/c2mt20055d
W. Y. Feng, M. Wang, M. Guan, Y. Hui, J. W. Shi, B. Wang, M. Zhu, Y. L. Zhao and Z. F. Chai,
Mercury speciation and mercury-binding protein study by HPLC-ICP-MS on the estimation of mercury toxicity between maternal and infant rats, J. Anal. At. Spectrom., 26 (2011) 156–164.
DOI: 10.1039/c0ja00111b
R. Janzen, M. Schwarzer,
M. Sperling, M. Vogel,
T. Schwerdtle,
U. Karst,
Adduct formation of Thimerosal with human and rat hemoglobin: a study using liquid chromatography coupled to electrospray time-of-flight mass spectrometry (LC/ESI-TOF-MS), Metallomics, 3 (2011) 847–852.
DOI: 10.1039/c1mt00043h
S. Trümpler, W. Lohmann,
B. Meermann,
W. Buscher,
M. Sperling,
U. Karst,
Interaction of thimerosal with proteins—ethylmercury adduct formation of human serum albumin and b-lactoglobulin A, Metallomics, 1 (2009) 87–91.
DOI: 10.1039/b815978e E.M. Krupp
E.M. Krupp, B.F. Milne,
A. Mestrot, A.A. Meharg,
J. Feldmann,
Investigation into mercury bound to biothiols: structural identification using ESI–ion-trap MS and introduction of a method for their HPLC separation with simultaneous detection by ICP-MS and ESI-MS, Anal. Bioanal. Chem., 390 (2008) 1753–1764.
DOI 10.1007/s00216-008-1927-x
J.P.K. Rooney,
The role of thiols, dithiols, nutritional factors and interacting ligands in the toxicology of mercury, Toxicology, 234 (2007) 145–156.
DOI: 10.1016/j.tox.2007.02.016
D.L. Rabenstein, A.A. Isab,
A proton nuclear magnetic resonance study of the interaction of mercury with intact human erythrocytes, Biochim. Biophys. Acta, 721 (1982) 371-384.
DOI: 10.1016/0167-4889(82)90092-1
S.C. Fang, E. Fallin,
The binding of various mercurial compounds to serum proteins, Bull. Environ. Contam. Toxicol., 15 (1976) 110-117.
DOI: 10.1007/BF01686202 Related EVISA Resources
Related EVISA Resources
 Brief summary: ICP-MS: A versatile detection system for speciation analysis
Brief summary: ICP-MS: A versatile detection system for speciation analysis Brief summary: LC-ICP-MS: The most often used hyphenated system for speciation analysis
Brief summary: LC-ICP-MS: The most often used hyphenated system for speciation analysis  Brief summary: ESI-MS: The tool for the identification of species
Brief summary: ESI-MS: The tool for the identification of species Link Database: Toxicity of Organo-mercury compounds
Link Database: Toxicity of Organo-mercury compounds  Link Database: Mercury exposure through the diet
Link Database: Mercury exposure through the diet  Link Database: Environmental cycling of methylmercury
Link Database: Environmental cycling of methylmercury Link Database: Environmental cycling of inorganic mercury
Link Database: Environmental cycling of inorganic mercury Link Database: Environmental pollution of methylmercury
Link Database: Environmental pollution of methylmercury Link Database: Environmental pollution of inorganic mercury
Link Database: Environmental pollution of inorganic mercury Link Database: Toxicity of mercury
Link Database: Toxicity of mercury Related EVISA News
Related EVISA News
 February 3, 2015: Mercury levels in Pacific yellowfin tuna increasing
February 3, 2015: Mercury levels in Pacific yellowfin tuna increasing September 2, 2014: Man is significantly contaminating oceans with mercury
September 2, 2014: Man is significantly contaminating oceans with mercury  May 3, 2013: Standard methods for mercury speciation analysis
May 3, 2013: Standard methods for mercury speciation analysis June 17, 2012: Factors Affecting Methylmercury Accumulation in the Food Chain
June 17, 2012: Factors Affecting Methylmercury Accumulation in the Food Chain August 16, 2010: Methylmercury: What have we learned from Minamata Bay?
August 16, 2010: Methylmercury: What have we learned from Minamata Bay? June 28, 2010: New Study Examines Why Mercury is More Dangerous in Oceans
June 28, 2010: New Study Examines Why Mercury is More Dangerous in Oceans August 21, 2009: USGS Study Reveals Mercury Contamination in Fish Nationwide
August 21, 2009: USGS Study Reveals Mercury Contamination in Fish Nationwide May 5, 2009: Ocean mercury on the rise
May 5, 2009: Ocean mercury on the rise February 11, 2009: Mercury in Fish is a Global Health Concern
February 11, 2009: Mercury in Fish is a Global Health Concern October 30, 2008: Precautionary approach to methylmercury needed
October 30, 2008: Precautionary approach to methylmercury needed March 11, 2007: Methylmercury contamination of fish warrants worldwide public warning
March 11, 2007: Methylmercury contamination of fish warrants worldwide public warning October 9, 2006: Linking atmospheric mercury to methylmercury in fish
October 9, 2006: Linking atmospheric mercury to methylmercury in fish September 23, 2006: Report Finds Mercury Contamination Permeates Wildlife Systems
September 23, 2006: Report Finds Mercury Contamination Permeates Wildlife Systems August 16, 2006: Mercury pollution threatens health worldwide, scientists say
August 16, 2006: Mercury pollution threatens health worldwide, scientists saylast time modified: January 15, 2025